Final Report
Cyanobacteria Detection And
Species Classification
By
Image Analysis
Cyanobacteria Risk Assessment for Lake Biwa (CRAB) Project
Dr Ross F. Walker
Lake Biwa Research Institute,
1-10 Uchidehama, Otsu, Shiga, 520-0806
In the following work we describe the application of image processing and pattern recognition techniques to the area of cyanobacteria detection and classification. Specifically, we target the species Microcystis sp. for detection and classification from among several other cyanobacteria species endemic to Lake Biwa: Anabaena flos-aquae, A. smithii, A. planctonica, and A. ucrainica. High-resolution microscope images containing a mix of the above species and other non-algal objects are analysed, and any detected objects are removed from the image further analysis. Following image enhancement, object properties are measured and compared to a previously compiled database of species characteristics. Classification of an object as belonging to class membership 'Microcystis' or 'other' is performed using parametric statistical methods. Leave-One-Out classification results suggest an error rate of approximately 2.3%.
Abstract
*Introduction
*Cyanobacteria Image Analysis
*System Hardware
*Image Processing Methodology
*Non-uniform illumination correction
. *Object segmentation
*Focus check
*Object feature extraction
*Feature selection
*Classification
*Results
*Classification Results
*Discussion and Conclusions
*Bibliography
*Appendix 1
*List of Features Extracted from Image Objects
*Appendix 2
*Author's Address
*
The increasing occurrence of algal bloom contamination in both lakes and sea serves as a worrying indicator of increasing environmental stress on water ecosystems. Such blooms have the potential to become a serious issue in terms of a government's ability to supply drinking water that meets national health standards, not to mention the devastating effects such blooms can have on the environment. Lake Biwa, Japan's largest lake and source of drinking water for over 14 million people, has experienced such blooms with increasing frequency over the last decade. The majority of these blooms are caused by 3 endemic species of cyanobacteria Microcystis sp., Anabaena sp., and Planktothrix sp.
Monitoring of water supplies (for the presence or absence of targeted species) usually involves the manual analysis of water samples by trained experts a very time-consuming and, therefore, expensive operation. Recent advancements in computer performance and image analysis now allow the possibility of assisted or 'adjunct' screening of water samples by image processing and pattern recognition systems. However, this area of research has seen minimal progress over the last decade, with very little published literature. Apart from THIEL 1994, there appears to be no other major published works on this subject, and very few active researchers working on this significant problem.
In this correspondence, we detail the implementation and evaluation of an image processing system for automatically detecting and classifying cyanobacteria taxa, and report classification results. We firstly introduce the topic of cyanobacteria image analysis and discuss several problems that make such analysis inherently difficult. We then discuss the hardware, software, and general image processing methodology that has resulted from the approximately one-year life of this project. Finally, we present the significant results of our research and discuss relevant points that need further investigation.
While being very similar to other forms of image analysis, cyanobacteria classification presents the image analyst with its own inherent difficulties. For example, the types and numbers of objects (bacteria, zooplankton, debris etc.) that may be present in any one sample of lake water is both unknown and effectively unlimited. Also, intra-species variation of characteristics (such as size and colony shape) can be large, and is often seasonally dependent. Furthermore, the bulk size of two targeted species may be an order of magnitude or more apart, making decisions such as image magnification setting a difficult choice. The dynamic and variable nature of the lake microcosm thus creates a formidable challenge to the design of a robust pattern recognition (P.R.) system with the ideal characteristics of high accuracy but with wide generalisation ability.
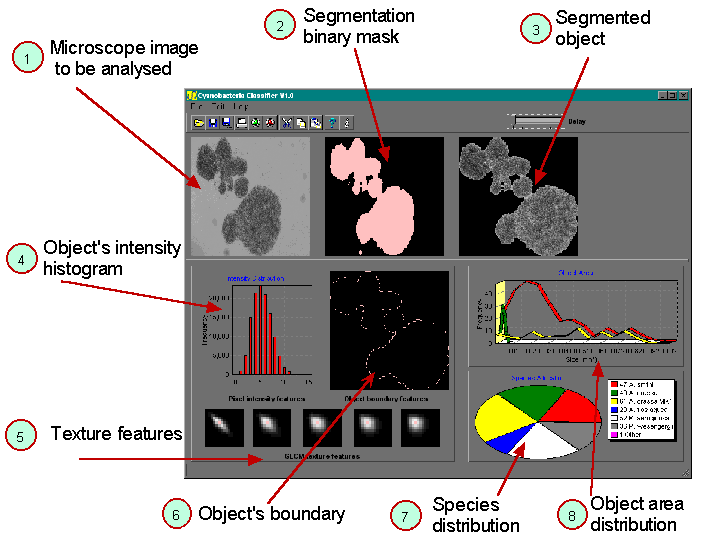
The work we present here is the second stage of a preliminary study originally published in WALKER, TSUJIMURA, KUMAGAI, 1998. The image processing system has been totally re-designed and built from scratch, and following training, can now operate completely autonomously. Figure 2 and Figure 3 show the system, consisting of purpose-built light microscope with water sample feed system, high-resolution digital imaging camera, image server computer, and several image processing computers. Sample water is passed as a continuous stream under the microscope's objective. Digital images of this water are sequentially captured by the image server, which then transfers each image to an available image processing computer. Objects contained within any image are analysed, and the analysis results returned to the server for display see Figure 1.
For the purposes of this study, we classified image objects as being either from the class 'Microcystis' or from the class 'other'. By the class 'other' we mean all image objects that are not from the genus Microcystis. Such objects include other cyanobacteria species such as Anabaena, zooplankton, weed, sediment, air bubbles, etc.

Figure 1 The image analysis process. (1) Microscope image of water sample is transferred to computer for analysis; (2) Segmentation mask is determined; (3) Object to be classified is removed from the surrounding image background; (4-6) object characteristics are quantitatively measured; (7) Relative distribution of species numbers found in the water sample; (8) Distribution of object areas.
Imaging hardware consists of a custom-built optical microscope, high-resolution greyscale digital camera, and digital frame grabber see Figure 2. Image magnification and focus are controlled via a single remote x-y joystick. However, once appropriate magnification is chosen, it is not varied. Sample water is passed through a water channel cell see Figure 3. This was manufactured with a channel depth of 0.8mm, to allow for large objects to flow through the cell channel without clogging. However, at the magnification used, focal depth of the microscope optics was approximately 0.2mm. As we have no control over where in the 0.8mm channel depth objects will appear, poorly focused objects can and do occur. We handle this problem by measuring focal accuracy of each image object prior to processing, and only classifying those objects with adequate focus. We chose to image the lower 0.2mm of the 0.8mm channel depth, as gravity helps to shift floating objects to this level.
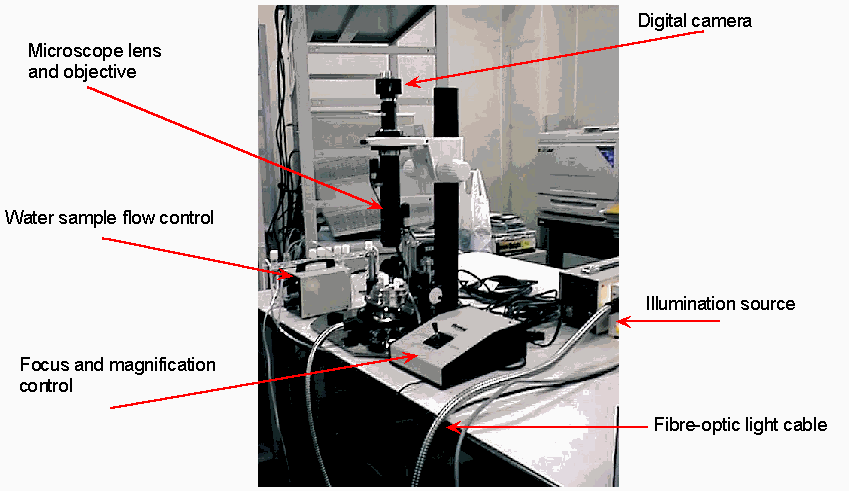
Figure 2 Microscope and high-resolution digital imaging system.
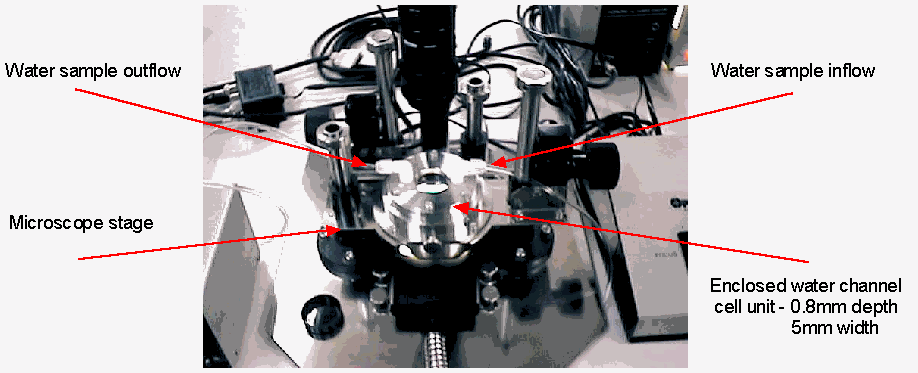
Figure 3 The microscope stage and water sample channel, shown in greater detail.
Non-uniform illumination correction.
Digital images received for analysis are pre-processed to reduce the effects of non-uniform illumination. Such an undesirable characteristic, which represents a non-linear transformation of the true image data, can present a severe challenge to subsequent image processing algorithms such as segmentation. In the present system, non-uniform illumination is crudely corrected by cropping the image borders. However, in a soon to be implemented step, the microscope's illumination transfer function will be measured and used to correct the images. Figure 4 shows the illumination response of the microscope currently in use.

Figure 4 Illumination response of the microscope (in false colour). Notice the strong non-uniform characteristic that can lead to classification inaccuracies if not corrected.
Objects within each image are separated from the image background via the process called segmentation. By object, we mean any body (group of image pixels) that appears darker than the image background. This can include cyanobacteria, as well as dirt, non-bacteria species, weed, etc. We achieve segmentation by forming a binary segmentation mask, and overlaying this mask on the original grey-scale image. Areas of the image that show through the mask (the objects) are then removed from the image and processed.
Initially, a rough mask is formed by a simple thresholding of the image at an intensity level determined by the intensity statistics of the image. This mask is then further processed to remove small imaging 'noise' particles or other objects smaller in size than the minimum expected size of the targeted cyanobacteria species. Finally, mask edge pixels are smoothed to form more uniform object boundaries. These steps are performed via morphological image processing algorithms operations based on mathematical morphology (SERRA 1982, VINCENT & BEUCHER, 1989). Figure 5 details the segmentation of a typical water sample image.
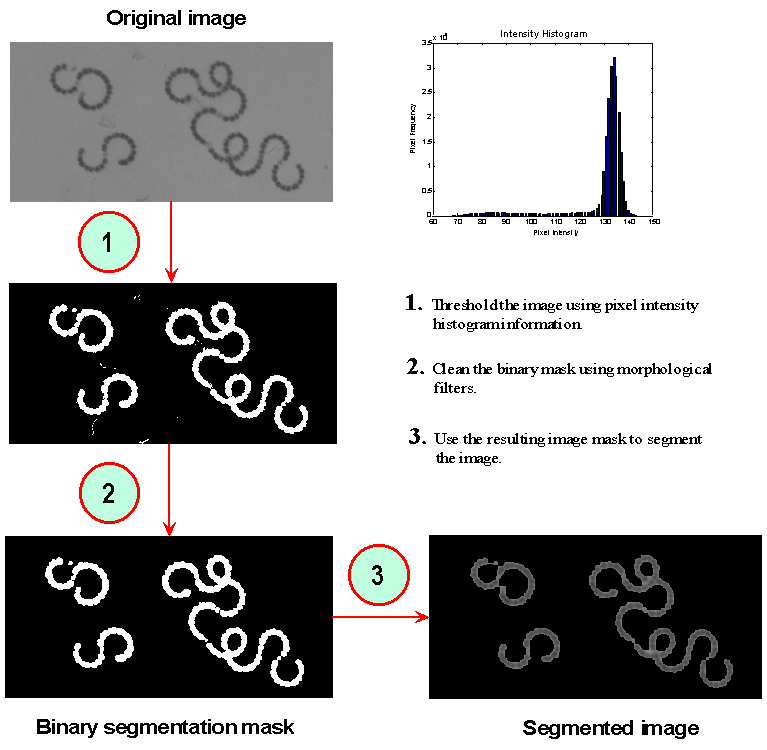
Figure 5 Segmentation of a typical water sample image.
Segmented objects are individually checked to ensure they possess an adequate level of focus. This step is vitally important. Because the water source is a 3-dimmensional column, there is no control over whether an object will fall within the in-focus portion of the microscope's view. As a consequence, there can be great variability in the focal accuracy of objects appearing in the microscope's field of view. Objects with adequate focus are subsequently processed and classified. Objects that do not achieve a minimum average focus limit are analysed to measure simple characteristics such as area and shape, but are not subsequently classified. This is because the defocus effect adversely influences many of the statistical properties of the image, and thus may have a strong negative influence on classification accuracy.
Focal quality is measured by isolating the middle to high spatial frequency components of the object image, and averaging the remaining power spectrum across one image dimension (OLIVA, BRAVO-ZANOGUERA & PRICE, 1998),
![]()
where ![]() is the focal quality measure for greyscale image
is the focal quality measure for greyscale image ![]() of spatial domain
of spatial domain ![]() pixels, and
pixels, and ![]() is the spatial domain response of the 1-D highpass filter kernel. This is a widely used technique and is computationally light.
is the spatial domain response of the 1-D highpass filter kernel. This is a widely used technique and is computationally light.
Image objects of adequate focal quality and exceeding minimum size requirements are sequentially extracted from the image for further processing.
To accurately classify an object into one of several classes, i.e., Microcystis, Anabaena, etc., it is necessary to quantitatively measure characteristics of the object that may indicate its class membership. For example, the characteristic 'area' is a good discriminator of class membership when classifying Microcystis and Anabaena cyanobacteria, as these two geneses differ substantially in size Microcystis usually being an order of magnitude larger in area. In pattern recognition terms, we call these characteristics 'features', and the process of measuring an object's characteristics as 'feature extraction'.
We measure a total of 123 object features including morphometric properties, object boundary shape properties, frequency domain properties, and second-order statistical properties (see Table 1). A full list of features can be found in Appendix 1.
|
Feature type |
Examples |
Number measured |
|
Morphometric |
area, circularity |
4 |
|
Boundary shape |
curvature properties |
5 |
|
Frequency domain |
Fourier components of boundary |
14 |
|
Second-order statistical properties |
Grey level co-occurrence matrix features |
100 |
Table 1 Types of features extracted from each image object.
Without a-priori knowledge, it is difficult to know which of the 123 object features will be useful for distinguishing between cyanobacteria species. By the term 'useful' we mean features whose statistical properties (mean and variance/co-variance etc.) differ between the various classes of data we are trying to classify. We call such features 'discriminatory', in that they can be used to discriminate between classes of data (see Figure 6).

Figure 6 An example that illustrates discriminatory power. The feature on the left possesses low discriminatory power, while the one on the right possesses high discriminatory power. Features whose class-conditioned distributions overlap the least will have greater discriminatory power.
Selecting a subset of discriminatory features from a larger set is called 'feature selection'. The process is arguably one of the most important steps in pattern recognition. Generally, there will exist a high dimensional feature space, with a limited number of data samples to accurately characterise the class distributions within this space. By removing redundant features that do not discriminate between classes, we can better represent this now lower-dimensional space, allowing us to design a more robust classifier.
To find an optimal feature subset, we used a feature selection process called sequential forward-selection/backward-elimination (HAND 1981). Specifically, our algorithm adds two new features and then removes one feature, thus capturing feature pairs that possess higher-order discriminatory power. KITTLER 1978 reported that this method almost always gave optimal results and computationally was comparable to less optimal approaches. Using this method, we found that a total of five features from the original set of 123 possessed significant discriminatory power to classify the object data as being either from the classes Microcystis or Other.
We performed classification using a general Bayes decision function for assumed Gaussian feature distributions with unequal variance-covariance matrices (GONZALEZ AND WOODS, 1993). The resulting decision surface (where ![]() ) is of hyperquadric form:
) is of hyperquadric form:
![]()
where ![]() is the feature vector of the object to be classified,
is the feature vector of the object to be classified, ![]() represents the discriminant measure for
represents the discriminant measure for ![]() ,
, ![]() is the a-priori probability of class
is the a-priori probability of class ![]() , and
, and ![]() and
and ![]() are the variance-covariance matrix and mean vector respectively for class i data (determined from a database of objects from known class). Because we are only classifying image objects into 2 classes Microcystis or other only one classification stage is required. Our previous work (WALKER, TSUJIMURA, & KUMAGAI, 1998) has shown that the extension to classifying multiple species is both straightforward and relatively accurate when abundant images of high quality are used.
are the variance-covariance matrix and mean vector respectively for class i data (determined from a database of objects from known class). Because we are only classifying image objects into 2 classes Microcystis or other only one classification stage is required. Our previous work (WALKER, TSUJIMURA, & KUMAGAI, 1998) has shown that the extension to classifying multiple species is both straightforward and relatively accurate when abundant images of high quality are used.
A total of 1529 image objects were extracted from among the 1468 images of the five cyanobacteria species. Of these, 1348 were found to have adequate focus and were subsequently classified into the two classes of Microcystis or Other.
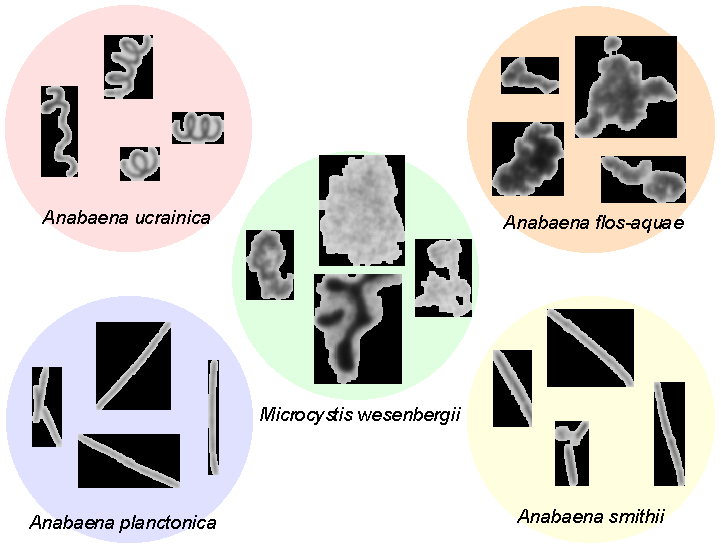
Figure 7 Examples of individual cyanobacteria that were extracted from high-resolution images, captured using the hardware described in this document.
Table 2 is a confusion matrix of general classification results. Total real error was measured to be 2.3% using the leave-one-out technique (WEISS & KULIKOWSKI, 1991).
Classified As Species:
|
Species |
Microcystis |
Other |
|
Microcystis |
247 |
19 |
|
Other |
12 |
1070 |
Table 2 Confusion matrix of classification results.
In Table 3 we present the classification results in more detail by separating the class 'Other' into its four constituent species. We notice that the species A. flos-aquae has a higher rate of misclassification than the remaining classes. We suspect this is because A. flos-aquae, while being of spiral shape like A. planctonica, forms itself in very tight, condensed spirals see Figure 7. As a result, the physical shapes of some A. flos-aquae specimens had attributes similar to the dense colonies of Microcystis, and were subsequently misclassified. However, the 9 misclassified samples represents an error rate of only 2.7%, which we feel is sufficiently low.
Classified As Species:
|
Species |
Microcystis |
Other |
|
M. wesenbergii |
247 |
19 |
|
A. flos-aquae |
9 |
322 |
|
A. planctonica |
0 |
102 |
|
A. smithii |
2 |
336 |
|
A. ucrainica |
1 |
310 |
Table 3 Confusion matrix of classification results, showing species-specific results.
The work presented in this correspondence represents the latest phase of research that began in early 1998, and was documented in two publications (WALKER, TSUJIMURA, & KUMAGAI, 1998-1; WALKER, TSUJIMURA, & KUMAGAI, 1998-2). The initial intent of this research was to determine whether it was feasible to automatically classify cyanobacteria species contained in lake water samples, using image processing and pattern recognition techniques. If proven successful, such a system could be used to augment work currently done by trained bacteria experts, with significant benefits:
A number of questions remain to be answered by future research. Firstly, the bacteria samples used throughout this work were laboratory specimens. This was necessary due to the lack of available natural (lake) species at the times when image databases were compiled. As such, the intra-species variability which exists in lake species did not exist in the lab specimens. Moreover, the range of possible non-targeted species (zooplankton, weed, sediment, etc.) is far greater in the lake environment. As such, we need to stress that the results presented in this report are optimistic, and will without doubt degrade when natural lake water samples are used. The extent to which the results degrade will be the focus of our continuing research. We are looking forward to the summer months when most cyanobacteria species will be in abundance.
Despite the above reservations, we feel that the significantly low error rates reported in this and previous reports indicates that the automatic classification of cyanobacteria is indeed a feasible and relatively accurate alternative to manual classification.
OLIVA, Michael A., BRAVO-ZANOGUERA, Miguel, & PRICE, Jeffrey H., 1998: Autofocus for Phase-Contrast Microscopy: Investigation of Causes of Non-Unimodality, SPIE Proc. Optical Diagnostics Biological Fluids Advanced Techniques Analytical Cytology , pp3260, 1998
GONZALEZ, R.C. & WOODS, R.E., 1993: Digital Image Processing, Addison-Wesley, USA.
HAND, D.J., 1981: Discrimination and Classification, Wiley, USA.
KITTLER, J., 1978: Feature Set Search Algorithms. In C. H. Chen, editor, Pattern Recognition and Signal Processing. Sijthoff and Noordhoff, The Neatherlands.
KUMAGAI, Michio, 1996: Study on Climate-Induced Water Quality Change in the South Basin of Lake Biwa, Lake Biwa Research Institute Bulletin, No. 13, p16-19.
SERRA, J., 1982: Image Analysis and Mathematical Morphology, Academic Press, London.
THIEL, S.U., 1994: The Use of Image Processing Techniques for the Automated Detection of Blue-Green Algae. Ph.D. dissertation, University of Glamorgan.
VINCENT, L., & BEUCHER, S., 1989: The Morphological Approach to Segmentation: An Introduction, in Mathematische Morphologie und Digitale Bildverarbeitung, München, 25-27 September, 1989.
WALKER, R.F., 1997: Adaptive Multi-Scale Texture Analysis with Application to Automated Cytology. Ph.D. dissertation, University of Queensland, Australia.
WALKER, Ross F., TSUJIMURA, Shigeo, & KUMAGAI, Michio, 1998-1: Automatic Monitoring of Six Cyanobacterial Taxa From Lake Biwa By Image Processing, SIL '98, the 27th Congress of the International Association of Limnology, 9th-14th August, 1998, Dublin, Ireland.
WALKER, Ross F., TSUJIMURA, Shigeo, & KUMAGAI, Michio, 1998-2: Automatic Hierarchical Classification of Cyanobacteria by Image Processing and Pattern Recognition, Proceedings of the 63rd Annual Meeting of the Japanese Society of Limnology, 20th-23rd September, 1998, Matsumoto, Japan, pp126
WEISS, S.M. & KULIKOWSKI, C.A., 1991, Computer Systems that Learn: Classification and Prediction Methods from Statistics, Neural Networks, Machine Learning, and Expert Systems, Morgan Kaufmann, San Mateo.
List of Features Extracted from Image Objects
For a complete description of the twenty texture features, see WALKER, 1997.
Dr Ross F. Walker, Lake Biwa Research Institute, 1-10 Uchide-hama, Otsu City, Shiga Japan, 520-0806.
Email: walker@lbri.go.jp Web: https://members.tripod.com/~Dr_Ross_F_Walker