
BIONT
Modeling Program for the Complex Biochemical Systems
 |
BIONT is the program for simulation and
analysis of biochemical kinetics, membrane transport kinetics and volume
changes in complex multicompartment systems like Mitochondrial
Permeability Transition system. BIONT is written in MatLab 5.11
environment, which provides excellent tools for handling large data
matrixes and good possibilities for graphical data representation. |
We prefer to write BIONT, instead of using other available
biochemical simulation programs, due to necessity to manage some distinctive
properties of compiled MPT model, not readily handled simultaneously by the most
of convenient programs:
 | Simultaneous electrogenic membrane transport processes and membrane potential-dependent
processes (e.g. ion transporters, respiratory chain, and ANT)
 | Varying compartments' volumes (e.g. mitochondrial matrix,
intermembrane space)
 | Multicompartment and multimembrane system |
 | Various reactions' kinetics, often not coincided with widespread types.
 | Very large number of first-order differential equations (in perspective
- arbitrary first-order differential equations) and variables - about
several hundreds and more. |
| | |
Calculation principles used in BIONT
Uniform additive variables
We use uniform additive variables which could represent any of the following
parameters in the similar way:
 | Substances' concentrations
 | Enzymes' concentrations
 | Transmembrane charge
 | Media's volumes |
| | |
First order differential equations
 | We use for calculations uniform 4-parameter first order non-linear differential equations. (4
parameters: forward rate constant, reverse rate constant, charge
transferred, electrogenic reaction type)
(In perspective - arbitrary first order differential equations, build-in
standard kinetics types)
 | During integration program performs continuous estimation of membrane(s)' potential(s) and compartments' volumes
and, according to the values corrects reactions' rates and substances'
concentrations.
 | (In perspective - continuous estimation of osmolarity, ionic force and pH control: for
...-dependent reactions)
 | Modified Rosenbrock formula of order 2 for stiff systems was used for
numerical integration
(In perspective - various integration methods)
 | Calculation algorithm can use adaptive time step size, according to the fastest reaction
rate |
| | | |
Program features
Models handling
 | Multiple substances number (up to 10000 - limited by RAM mainly)
 | Multiple compartments
 | Multiple charged membranes
 | Varying compartments' volumes
 | Varying membranes' charges
 | (Sub)models creation, changing (add/remove substances and/or reactions),
extraction, combining, deletion, saving, loading
 | Direct table values edition
 | Conveniences complex (e.g. substances-reactions-calculation-plotting
synchronization, automatic reaction rate/equilibrium constant/rate value
preview) |
| | | | | | |
Behavior calculation
 | Adaptive/fixed time periods
 | Automatic curve groups calculation at some parameter change (e.g. number
of concentration/time curves at different reaction rates)
 | Intermediate steps saving (on HDD) and recall
 | Metabolic control analysis suit
 | Control coefficients
 | Elasticity coefficients |
|
|
| | |
Visualization
 | Multiple plots
 | Using calculated values buffer (no need to recalculate for replotting)
 | Parameter/time curves
 | Parameter/parameter curves
 | Built-in parameters' functions set (ln, statistical, special reaction rate
functions - dG, partial rate etc.)
 | Custom functions calculation and plotting |
| | | | |
Parameters optimization
 | Various optimization criteria
 | any parameter's arbitrary or built-in functions' value/minimum/maximum
(e.g. reaction rate should be 5 nmol/min, or substance concentration
should be as maximal as possible)
 | interparameter dependencies' curves shape (e.g. S-type dependence
according to experimental curve shape)
 | multiple simultaneous optimization criteria (e.g. reaction rate should
be 5 nmol/min and at the same time some substance concentration should
be as maximal as possible)
 | selective multiple optimizable parameters (e.g. find out
concentrations' values giving maximum reaction rate)
 | synchronous/free parameters optimization (e.g. we have fixed
reactions' rates ratio but need to fit absolute values)
 | fixed/free optimization ranges |
| | | | |
 | Various optimization methods
 | Gradient
 | Multigradient
 | Simplex
 | Genetic
 | Random search |
| | | |
 | Optimization conveniences set |
| |
Statistical estimation
 | Statistical analysis suit
 | Dispersion of multiple model parameters
 | Normal/uniform dispersion
 | Dispersion range customizable for each parameter
 | Synchronous/free parameters dispersion |
| | |
 | Various statistical parameters estimation and appropriate graphical
representation tools
 | Average
 | Standard deviation
 | Correlation
 | Normality
 | Histogram
 | Asymmetry, Excess, Box plot, etc. |
| | | | |
 | Statistical estimation of any custom or built-in parameters' functions |
| |
Elements of Metabolic Control Analysis
 | Metabolic control analysis suit:
 | Elasticity coefficient estimation |
 | Control coefficient estimation |
 | Additional combined parameters |
|
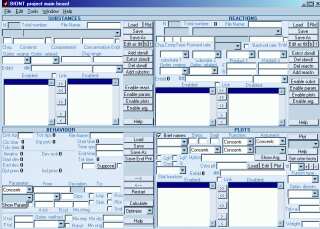
Program screenshots:
(Click to enlarge)
| Main program window: |
Plots window: |
 |
 |
Your comments or questions are welcome!
|



