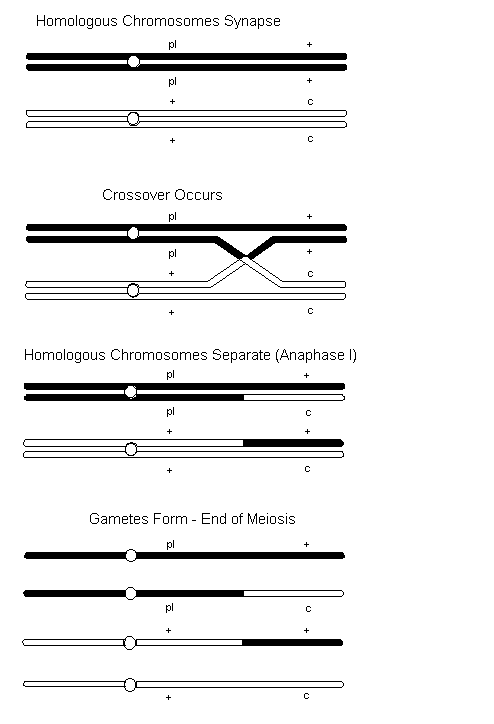
When two or more genes reside on the same chromosome, whether sex chromosome or autosome, they are said to be linked. Linked genes do not independently assort during gamete formation. However, linked genes do not always stay together because homologous non-sister chromatids may exchange segments of varying length with each other. This chromatid exchange, referred to as crossover, occurs during prophase I of meiosis when homologous chromosomes synapse. In the Figure below, the sex-linked genes for Pearl (pl) and Cinnamon (c) are used to demonstrate crossover. In this case, the male is heterozygous for both traits. You will also notice that the mutations are arranged such that they do not both occur on the same chromosome. This arrangement ([+,pl][c,+]) is referred to as repulsion. If both dominant (or wild-type) alleles are on one chromosome and both recessive (or mutants) are on the other the linkage relationship ([+,+][pl,c]) is referred to as coupling.

From the above example, the male can provide four different types of gametes to the fertilization process ([pl,+] [pl,c] [+,+] and [+,c]). If coupling was applied to the above example the type of gametes produced would be the same. The only difference between the two situations is the statistical outcome.
Crossover rate can be effected by sex, age, temperature, proximity of allele to the centromere, inversion and many other factors. The highest probability of crossover that could be seen between any two genes for any species is no more than 50%. If crossover was to occur at its maximum frequency (~50%) then the alleles would appear to be independently assorting.
With sex-linked traits, crossover only occurs in males because only males have a homologous set of sex chromosomes. Currently, I am unaware of any linked mutations on the autosomes. If such a condition was to arise, then crossover could be observed and it would be at the same frequency for both sexes.
The physical point at which chromosomes exchange during crossover is referred to as a chiasma (plural chiasmata). Each type of chromosome within a species has a characteristic (or average) number of chiasmata. The frequency with which a chiasma occurs between any two genes also has a characteristic or average probability. The further apart two genes are located on the chromosome, the greater the opportunity for a chiasma to occur between them. The probability of chiasma occurring between two linked genes is calculated as follows:
| Chiasma % = 2(crossover %) | or | Crossover % = 1/2(chiasma %) |
Therefore, if you observe 30% of the crossover genetic linkage in your offspring, such as [pl, c] and [+, +] in the above example, then the Chiasma frequency is 60%. Chiasma frequency is twice the crossover frequency because when a chiasma forms between two genes, only half of the meiotic products will be of the crossover type. In other words, chiasma is the probability that chromosomes (i.e. two chromatids) will exchange between a synapsed pair of chromosomes (bivalent) containing four chromatids - also referred to as a tetrad.
The chromosomes that are not involved in the crossover are referred to as non-crossover or parental types. Those involved with the crossover are referred to as recombinant or crossover types.
In the above example they are as follows:| [pl , +] [+ , c] |  | Parental |
| [pl , c] [+ , +] |  | Recombinant |
Next, I will discuss application of crossover to the Punnett square. When applying sex-linked genes to the Punnett square, the crossover gametes produced by the male should be included. Again, the female cannot produce any crossover gametes for sex-linked genes. I will use the example above with a Gray Male heterozygous for Pearl and Cinnamon in repulsion phase (X+,plXc,+) crossed to a Gray Female (X+,+Y). See Figure 18.
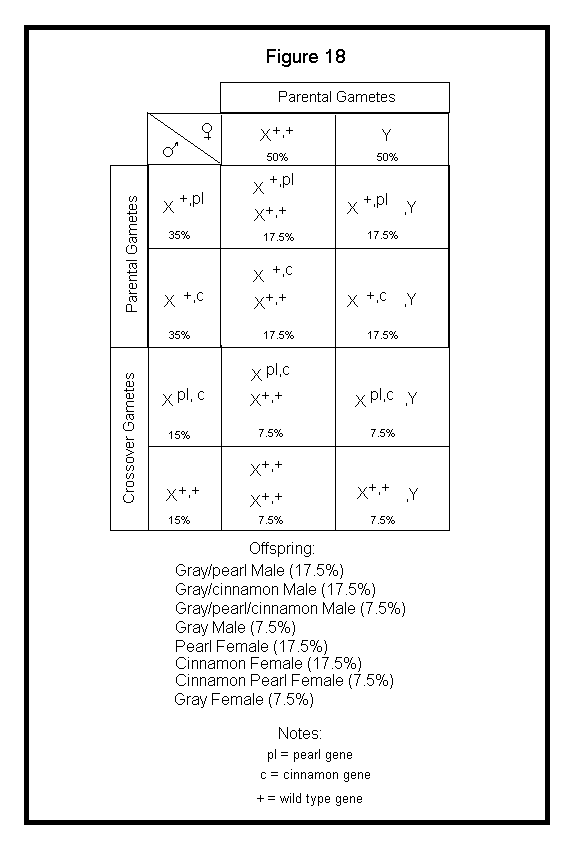
In the above case, I arbitrarily assumed a 30% crossover rate for the two loci. I have been unable to locate genetics information on cockatiels that provides crossover frequencies for the various linked mutations. I suspect we are still in the data collecting phase.
Let us assume we have a male heterozygous for four sex-linked traits. This would then present three various regions for crossover to be observed. See Figure 19.

| Xa,b,c,d X+,+,+,+ |  | Parental |
| Xa,+,+,+ X+,b,c,d Xa,b,+,+ Xa,b,c,+ X+,+,+,d |  | Single Crossover Recombinants |
The maximum frequency of recombinant gametes for any two loci is 50%, because this would represent 100% single chiasma frequency. Gene distances are expressed as map units. One map unit is equivalent to 1% recombinant progeny. If the a-b distance is 50 map units, b-c is 30 map units, and c-d is 40 map units, the distance a-d would be 120 map units. But in a cross involving segregration of only the two most distant markers (a-d), the number of recombinant progeny would not be expected to exceed 50% because of mulitple crossover, some of which produce the equivalent of noncrossover.
When discussing more than two genes on a chromosome that experience crossover then you must also factor in two strand double crossovers and multiple even-numbered crossovers. The figure below shows the sequence of events involved in double crossover using a male heterozygous for three sex-linked traits.

| Parentals | Xa,b,c X+,+,+ | 72% |
| Single Crossover (b-c) | Xa,b,+ X+,+,c | 18% |
| Single Crossover (a-b) | X+,b,c Xa,+,+ | 8% |
| Double Crossover | Xa,+,c X+,b,+ | 2% |
Note that in the above example, the rate of double crossover (2%) has been subtracted from the total crossover frequency for a-b (10%-2%=8%) and b-c (20%-2%=18%) to derive the single crossover rates. Additionally, the following can be stated:
Double crossover has been studied in fruit flies and from these studies, it has been determined that double crossover usually does not occur between fruit fly genes less than 5 map units apart (1 map unit=1% crossover rate). In the above example, the distance between genes a and c is 30 map units (i.e., 30% total crossover rate including single and double crossovers = 30 map units).