Introduction
Distributed object technology is considered to be a revolution for software design in heterogeneous computing environments. Using this approach, an application is abstracted and divided into self-managing objects that can interoperate across heterogeneous networks and operating systems. An object can be thought of as a set of data and operations (or methods) that can be performed upon the data.
Since 1989, the Object Management Group (OMG) (URL: http://www.omg.org), a consortium of >700 member organizations, has been focused on specifying the architecture on which objects written by different vendors can interoperate across networks and operating systems. Its main product is the specification of the Common Object Request Broker Architecture (CORBA) (Siegel, 1996; OMG, 1997b). The CORBA specification describes a software bus, called an Object Request Broker (ORB), that provides an infrastructure on which a client can invoke the methods of server objects without the knowledge of where the server objects are located, how they are implemented, whether the object is currently activated and what communication mechanisms are used (Vinoski, 1997). Usually, a client only needs to know what types of operations an object can provide, i.e. the object's interface. Object interfaces are defined using OMG Interface Definition Language (IDL). IDL is a declarative language (not for programming) which forces interfaces to be defined separately from object implementation. Therefore, language independence, an important feature within heterogeneous systems, is supported (Vinoski, 1997). The ORB and the IDL in CORBA provide the basic mechanisms to support distributed object computing.
Based on the CORBA concepts mentioned above, the OMG proposed a reference architecture for distributed object computing, called the Object Management Architecture (OMA) (OMG, 1997a), shown in Figure 1. Object Services provide a set of basic services that are likely to be used by any object, e.g. a service which allows clients to find an object based upon names (Naming Service). Adopted OMG Object Services are collectively called CORBAservices. Common Facilities (sometimes called horizontal common facilities) provide services oriented towards domain-independent end user applications (not towards objects in Object Services), e.g. user interface management. Adopted OMG Common Facilities are collectively called CORBAfacilities. Domain Interfaces (sometimes called vertical common facilities) provide standardized facilities for specialized application domains, e.g. finance and health. Domain Interfaces will be grouped by application domain. The Application Interfaces are developed specifically for a given application.
In late 1994, a protocol for ORB interoperability called the Internet Inter-ORB Protocol (IIOP) was approved in the CORBA 2.0 specification, which allows software components developed using ORBs from different vendors to interoperate. IIOP runs on top of TCP/IP and is becoming the standard for communication among distributed objects running on the Internet and enterprise intranets. Leading vendors of Internet tools, like Netscape, have fully endorsed IIOP in their current and future products (Netscape Communications Corporation, 1997).
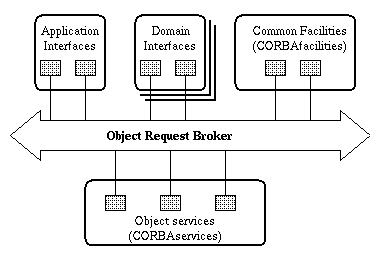
Figure 1. OMA reference model.
There is a growing interest in applying CORBA in the bioinformatics area. For example, the European Bioinformatics Institute (EBI) has recently developed a number of prototype CORBA systems for bioinformatics, including one that provides access to chromosome maps in a radiation hybrid database (URL: http://industry.ebi.ac.uk/~corba). There is also a mailing list called `BioObjects' (URL: http://sunny.ebi.ac.uk/BioObjects/) which aims at facilitating discussion, exchanging ideas and experiences. At a recent 2-day meeting-Objects in Bioinformatics, held at the EBI in June 1997-many participants showed a strong interest in using CORBA for a variety of biological applications. In August 1997, the OMG set up a domain special interest group: the `Life Sciences Research' group (URL: http://lsr.ebi.ac.uk). One of its main goals is to use the OMG technology adoption process to standardize interfaces for software tools, services, frameworks and components in life sciences research.
The Bioinformatics Group at the Roslin Institute is committed to providing animal genome mapping information services to the worldwide molecular biology community. We have developed a genome database (ArkDB) to handle genome mapping data for a single species (Archibald et al., 1996). The ArkDB model provides a generic structure for all classes of animal genome mapping data. Currently, we have a cluster of different species (e.g. pig and chicken) genome databases which have the same schema and are implemented using the Ingres Database Management System (DBMS). World Wide Web technology has been employed as a front-end for information retrieval and data editing. The Web front-ends are constructed using WebinTool, a software tool that provides a framework for Web to SQL database interface building and maintenance (Hu et al., 1996).
We have investigated CORBA as a means of providing a coherent framework for future applications, of enabling Java as a user interface technology, and as an infrastructure for interoperation between genome databases. Here we discuss these issues and describe the development of a prototype CORBA gene mapping system.
System and methods
CORBA is only a specification. To build up a CORBA compliant application, one usually requires a software package which provides an ORB and an IDL compiler. The ORB supplies a framework which facilitates object communications and the IDL compiler takes the IDL input and generates appropriate source code in a given programming language such as C++.
The first and most important task in developing a distributed object system is the system design, in which the objects and the relationships between them are modelled at a conceptual level. This model reflects how users perceive the system, e.g. how the data are classified, what operations are required upon the data. End users are usually required to be involved during this design stage. Based on the conceptual model, an IDL file is then designed. The IDL compiler compiles the IDL file to generate the client side stub code and the server side skeleton code. A stub is a mechanism that creates and issues requests on behalf of a client, while a skeleton delivers requests to the CORBA object implementation. Note that the compiler for the stub and the compiler for the skeleton are not necessarily the same, e.g. a compiler performing IDL to Java mapping to produce a Java stub, and a compiler for C++ mapping to produce a C++ skeleton. Finally, the object server code and client application code need to be written. Figure 2 gives an illustration of CORBA-based application development.
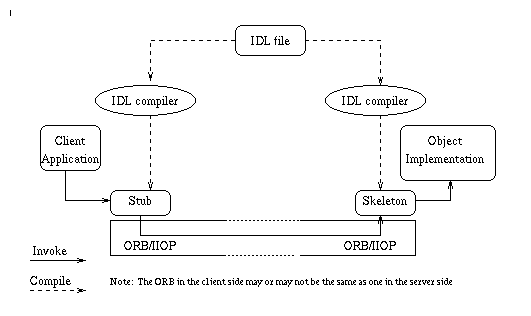
Figure 2. CORBA-based system development.
In the following, we discuss the development issues in the context of gene mapping systems.
Genome mapping data model design
The core of our system is the genome maps, each of which is a set of ordered and positioned loci (markers) along a chromosome. These maps need to be supported by experiments which are documented in external references. Based on this simple analysis, we designed four classes of objects in our genome mapping system: map, locus, experiment and reference.
1. Map. A map represents relative order and distance among loci along a chromosome.
2. Locus. A locus is a genomic segment whose position on a chromosome is defined experimentally.
3. Experiment. An experiment describes the locus or loci involved, the method used and the results observed.
4. Reference. A reference is an article, paper or book from which the data are drawn, and which provides a means of attributing the data to specific sources.
As a map represents order and distance among loci along a chromosome, the basic attributes of a map should include: the map name, the chromosome, a list of mapping points (or assignments) each of which references the locus and its position. The map concept can be further specialized, e.g. a cytogenetic (i.e. physical) map and a linkage (i.e. genetic) map. The basic attributes of a locus include its symbol, name and type. An experiment can be described by the method used and results observed. A reference can be defined by the title, authors, source, date and abstract.
The relationships among these objects are as follows.
1. A map's assignment list contains a set of loci, each of which can be detailed by the corresponding locus object. An assignment specified by a locus and its position along a chromosome should be supported by a specific experiment.
2. A locus may appear on one or more maps, and may be involved in several experiments.
3. An experiment may involve one or more loci and it must be documented by a reference.
4. A reference can record a set of experiments.
These object classifications and relationships are depicted in Figure 3 using the OMT(Object Modeling Technique)-like notation (Rumbaugh et al., 1991).
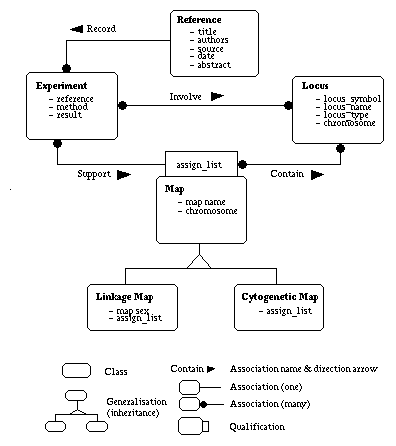
Figure 3. Conceptual model for genome mapping system.
Server side object implementation
Usually, the genome mapping data are stored in one or more databases. Many of the databases in use are relational and support the Structured Query Language (SQL) which is an industry standard. Thus, the first issue for server side development in genome mapping applications is the database connectivity.
The issue of database connectivity has been widely addressed in recent years. The industry standard Open Database Connectivity (ODBC) specification has been endorsed by many software vendors and there are a number of commercial ODBC products. Another widely accepted specification for database connectivity is the JDBC (Java DataBase Connectivity) API (Application Programming Interface) proposed by Sun Microsystems (Sun, 1997a), which defines a standard interface for Java programmers to access relational databases. Some leading database and tools vendors have already endorsed the JDBC API and are developing products using JDBC. For either ODBC or JDBC applications, the basic database connection is performed by a database driver. Database drivers are already available from a number of vendors, and they can be used for database connection in the CORBA object implementation. In addition, a developer may also construct his/her own database driver for database access. The basic functionalities of a driver include database opening and closing, and SQL query execution.
The major issue for the server side object implementation is to perform mappings between IDL objects and the data stored in relational databases. Baker (1996) summarized two methods to perform CORBA objects to relational data mappings. In the first method, the object creation and operations are coded, in part, using SQL. A (small) part of the frequently accessed data of an object may be cached to improve performance, but most of the data would be stored in the database and would be queried using SQL in the object's method code. The second method aims at storing the CORBA objects themselves in the database, i.e. a tight integration of the CORBA and the database. To achieve this, some tasks must be carried out, like mappings between the object definition and a relational schema, coding to store and retrieve data from the database for an object, and loading the object from the database to the memory. These tasks may be implemented manually or can be automated with the help of tools. The first method is quite straightforward, especially for those developers who have programming skills in building database applications, as there is no significant difference between those applications and a CORBA application. This is very applicable in the context of legacy database applications. The second method is more elegant, though some extra resources (tools) may be required. This method is more suitable for the creation of new applications, i.e. starting from a well-defined object schema, then mapping to a (new) relational schema.
Another issue to be considered with the object implementation is the choice of programming languages. Java and C++ are obvious candidates; they are both object oriented and, being popular languages, they are supported by most ORB vendors. Currently, Java is executed by a virtual machine and hence it is inferior to C++ in terms of performance. We decided that C++ was the best choice for our object implementation.
Client side user interface
As the use of the Internet continues to expand rapidly, applications are required to be instantly deployable and maintainable across a variety of different hardware and software platforms. Since Java has the features supporting these requirements, it has become the main programming language for Internet applications. Most recent CORBA products provide IDL to Java mappings and the OMG has been carrying out standardization of the IDL to Java mapping (OMG, 1996).
A main feature of Java is its applet facility. A Java applet is a small Java program that can be downloaded from a Web server to the requesting client (e.g. a Web browser) where it is interpreted by the client. The advantage of this mechanism is that installation, upgrading and cross-platform compatibility are handled automatically. There is a disadvantage in that download time can be significant if the client application is large and the network is slow.
Another factor that should be considered in developing a genome mapping system is the graphical user interface (GUI). Since important data such as maps are most easily interpreted when presented graphically, the provision of a graphical map viewer is essential. This map viewer takes the data from the relevant database as input and constructs a graphical representation of one or more maps. The map viewer also needs to provide functions such as zooming, and comparison between two or more maps.
The development of graphical interfaces to gene mapping data can be supported by some Java toolkits. For example, the Java's Abstract Windowing Toolkit (AWT) provides a set of classes that are used to create and organize windows, implement GUI components, such as buttons, labels, text fields, handle events, draw text and graphics, as well as image processing. A similar Java tool kit is Netscape's Windowing IFC (Internet Foundation Classes) which is a class library that contains objects and frameworks used to create applets and applications. The IFC is an extension of the AWT library and makes the task of creating Java applications simpler for a developer.
Implementation
A system prototype has been implemented using an evaluation copy of Visigenic's CORBA software: VisiBroker for C++ (Orbeline) 2.0 Release 1.51 and VisiBroker for Java 1.2. They provide CORBA applications development environments for C++ and Java, respectively. Our ArkDB genome databases serves as the data source. The implementation work includes IDL file design based on the conceptual model, server side object implementation which mainly deals with database connectivity and mapping from relational data to CORBA objects, and creating a client side graphical gene map viewer. The system architecture is illustrated in Figure 4.
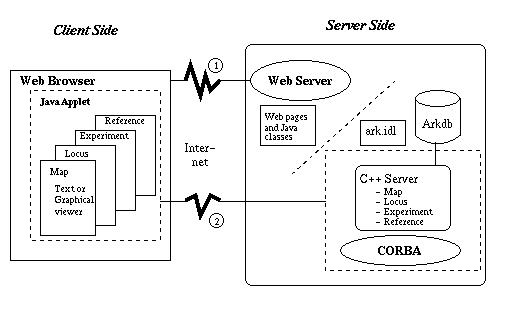
Figure 4. CORBA-based genome mapping system architecture. (1) Get a Web page and download the specified applet. (2) Interact with the CORBA server using the applet.
IDL file design
The IDL design is mainly based on the conceptual model which reflects data classifications and relationships; how to represent the model is a matter of investigation. Our initial IDL utilized a single interface with different operations for querying the database. Results were delivered via IDL structs. This has the advantage of simplicity, but does not take full advantage of CORBA IDLs and their potential for distributed object technology as it follows a more procedural model.
We then represented the conceptual model in IDL in a relatively straightforward way. An object class in the conceptual model is represented as an interface in the IDL specification, and the attributes of an object class are represented as attributes for the corresponding IDL interface. The relationships between classes are represented as appropriate methods or attributes in the relevant interfaces. While with this approach there are inherent efficiency disadvantages, it does allow exploitation of object-oriented methodology in the IDL design. The IDL specification is listed in the Appendix.
Server side implementation
The server is divided into two parts following the principle of modular programming: the database wrapper which performs database connections (like a JDBC or ODBC driver), and the CORBA object server which deals with mappings between CORBA objects and relational data.
1. Database wrapper. The wrapper has two layers: a core wrapper and an abstract wrapper. The core wrapper is the closest to the database and DBMS dependent. It provides three functions, described as follows (in C style):
/* open database */
int OpenDB(char* dbname,
char* dbuser,
char* password);
/* close database */
void CloseDB();
/* execute a query and get
results description */
ResultDesc* ExecQuery(char* query);
In this application where the Ingres DBMS is used, the core wrapper is implemented in C using the embedded SQL/C facility supported by the Ingres DBMS (a SQL/C++ facility is not provided at present) (Ingres Corporation, 1991).
The abstract wrapper uses C++ to specify some classes that implement basic functionalities for the database connection, SQL execution and results processing, such as:
(a) Connection represents a session of database connection and provides a method to perform a SQL execution to get a result set.
(b) ResultSet manages a result set generated by an SQL execution and provides methods to access the data.
The abstract wrapper is DBMS independent, but calls the functions provided by the core. The main purpose of the two-layer design is to keep DBMS-dependent code to a minimum in order to facilitate code portability.
2. CORBA object server. The code implements the objects specified by the IDL file using the VisiBroker's C++ application programming interface (API). It interacts with the abstract wrapper if database operations are needed. In this case, one or more relevant SQL statements are sent for execution and the results are returned to then be processed and assembled into appropriate CORBA objects.
When developing the object implementation, we were faced with design decisions concerning the logistics of database querying and object creation. On the one hand, we could fully utilize the power of relational databases to optimize SQL execution plans, and make extensive use of joins to fetch all the data in one single query. This has the disadvantage of fetching too much data which the client application does not necessarily require.
We chose to break down the operations into distinct queries for each interface. Where possible, only database internal identifiers are fetched and stored by the object implementation. This allows some queries to be delayed until the results are required. While this loses some of the power of database joins, it has the advantage that only data which are required are fetched. Overall, we have found that this leads to a more efficient object implementation. However, we anticipate a combination of these approaches, with large-scale joins being used by certain interface operations.
Client side implementation
The first client application we wrote to test the object implementation was a graphical map viewer. This is the most information-intensive part of our user interface, involving dynamic aggregation of large quantities of data (Mungall, 1996).
Our current public map viewer, Anubis, is a purely server-based application using CGI (the Common Gateway Interface). We are re-writing this in Java, transferring the processing burden to the client, where it belongs. An example of a Java-Anubis map display is shown in Figure 5. As static display does not do justice to an interactive viewer, we have mounted a demonstration available at http://www.ri.bbsrc.ac.uk/ark_corba/.
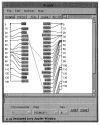
Figure 5. Anubis graphical map viewer.
With respect to CORBA, Anubis works as follows. Anubis is started with a particular default species. The first CORBA operation performed is a request for a list of map names for that species:
conn = dbserv.connect("");
map_names =
conn.getMapNames(species);
The user can select one of these names via a pop-up menu, along with chromosome and map sex. When the user has selected these parameters, and clicks on `Add map', the next CORBA operation is to fetch the map requested.
For instance, if the map type is a linkage (genetic) map:
Ark.LinkMap link_map =
Ark.LinkMap_var.narrow(
conn.getMap(
map_name, map_sex,
chromosome, species));
The list of map assignments can be accessed in order to draw the on-screen map:
Ark.PointAssign[] points =
link_map.map_point_list();
for (int i=0; i<points.length;
i++) {
Ark.Locus fragment =
points[i].locus;
float position =
points[i].position;
// now draw map
...
}
Because CORBA uses pass-by-reference, objects are never transferred, only values. This means that any graphically intensive application, such as a map viewer, must make repeated connections to the server as data must be frequently accessed, for instance, when zooming in on a map section.
To avoid this costly overhead, Anubis copies frequently accessed data into its own object space. This means Anubis has its own conception of a `locus' object, which is richer than the IDL representation. For example, the Java object contains a method for fetching all the on-screen representations of that locus.
Conclusion
As an open standard for distributed object computing, CORBA provides a framework for developing applications within distributed heterogeneous environments. CORBA specifies an ORB as the object communication infrastructure and the IDL as a language for object interface specification. We have examined some uses of CORBA for genome mapping information retrieval, and a prototype has been developed.
From our experience, the following benefits of using CORBA are evident.
1. Efficiency. CORBA provides a standard-based distributed object infrastructure, which frees developers from tedious low-level tasks, allowing them to focus on the problems at hand.
2. Flexibility. An object has a defined interface (described by the IDL). Changes to the implementation of the object do not affect clients and other objects so long as the object's interface remains the same.
3. Legacy integration. A legacy application need not be replaced. Instead, after being encapsulated in a thin `wrapper' to provide an appropriate interface, it can become a new component in the CORBA environment.
4. Interoperability. CORBA-compliant software objects are fully interoperable by using the IIOP, regardless of whether they use products from the same or different vendors.
Our next step will be to expand and improve the prototype. This will include further investigation of users' requirements, refining the IDL specifications (e.g. defining exceptions), enhancing the server performance using an object cache, improving the user interface utilizing the up-to-date Java technology and tools such as Java Foundation Classes (Sun, 1997b). In addition, we are going to look at any developments in domain interface standardization carried out by the Life Sciences Research Group. We believe that the interface standardization efforts will be beneficial for the genome mapping community and we are interested in making our contribution in this area.
Acknowledgements
The authors would like to thank the referees for their comments, which were very useful for the revision. We also appreciate Visigenic Software Inc. (http://www.visigenic.com) for granting the evaluation licence of its products. This work is funded by the UK Biotechnology and Biological Sciences Research Council.
Appendix. IDL for Arkdb
// File: ark.idl
// Description: IDL file for ARKdb
// Version: 1.2
//
module Ark {
interface Locus;
interface Reference;
interface Experiment;
interface Map;
interface CytoMap;
interface LinkMap;
typedef sequence
<Locus> LocusSeq;
typedef sequence <Map>
MapSeq;
typedef sequence <Experiment>
ExperimentSeq;
typedef sequence <string>
StringSeq;
// DBOBJ serves as a top level object
interface DBOBJ {
attribute string accession_no;
attribute string annotation;
};
interface Locus : DBOBJ {
attribute string locus_symbol;
attribute string locus_name;
attribute string locus_type;
attribute string chromosome;
attribute string species;
// get all maps on which the locus
// is present
MapSeqgetMaps();
// get all experiments in which
// the locus is involved
ExperimentSeqgetExperiments();
};
interface Map : DBOBJ {
attribute string map_name;
attribute string chromosome;
attribute Reference reference;
// get all loci appearing on
// the map
LocusSeqgetLoci();
};
struct RangeAssign {
Locus locus;
string arm_start;
string band_start;
string arm_end;
string band_end;
Experiment expt;
};
typedef sequence <RangeAssign>
RangeAssignSeq;
struct PointAssign {
Locus locus;
float position;
};
typedef sequence <PointAssign>
PointAssignSeq;
interface LinkMap: Map {
attribute string map_sex;
attribute PointAssignSeq
map_point_list;
};
interface CytoMap: Map {
attribute RangeAssignSeq
map_point_list;
};
interface Reference : DBOBJ {
struct author {
string surname;
string forenames;
};
attribute string title;
attribute AuthorSeq authors;
attribute string source;
attribute string date;
attribute string ref_abstract;
attribute string status;
// get all experiments recorded in
// the reference
ExperimentSeq getExperiments();
};
interface Experiment: DBOBJ {
attribute Reference reference;
attribute string method_desc;
attribute string result_desc;
// get all loci involved in the
// experiment
LocusSeq getLoci();
};
// A connection represents a session
// for an Ark service, which allows the
// client to interact with objects
//
interface Connection {
void close();
DBOBJ getDBOBJ(in string acc_no);
Map getMap(in string map_name,
in string map_sex,
in string chrom,
in string species);
MapSeq getMaps(in string chrom,
in string species);
StringSeq getMapNames(in string species);
};
// A DBserv provides an Ark service (as a
// starting object
//
interface DBserv {
Connection connect(in string name);
};
};